LET-99, a novel regulator of Galpha during spindle positioning in C. elegans embryos
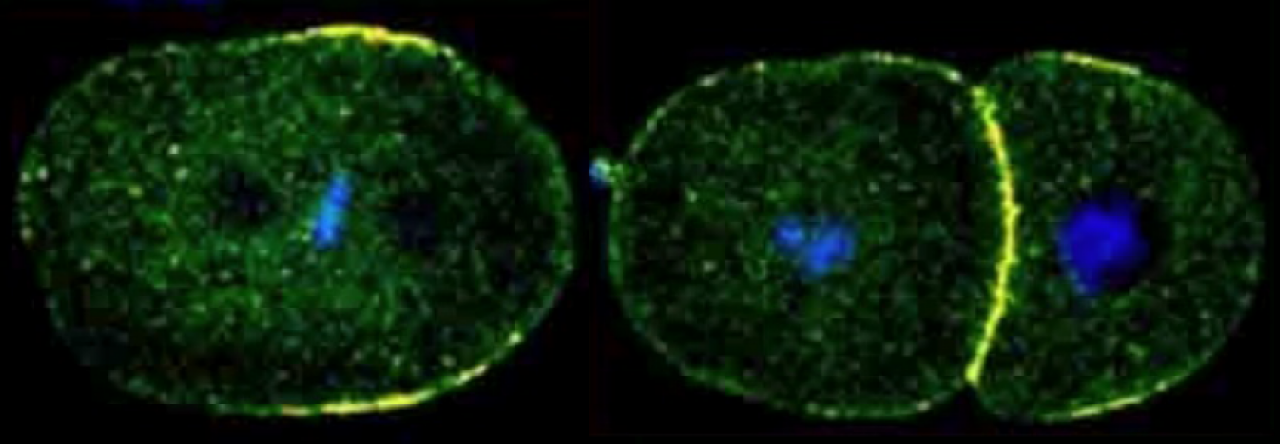
C. elegans embryos exhibit an invariant pattern of asymmetric and symmetric divisions. The conserved PAR polarity proteins the division of the asymmetrically dividing one-cell embryo, called P0, as well as its daughter P1. After fertilization, the PAR proteins become localized in opposite mutually exclusive domains of the cell (anterior PARs and posterior PARs in the C. elegans one-cell). The activities of the PAR proteins results in the polarization of cell fate determinants in the cytoplasm which is coordinated with the plane of division so that determinants are differentially segregated to the daughter cells. Division plane orientation depends on the position of the mitotic spindle and thus elucidating the molecular mechanisms that move nuclei and orient spindles are key to understanding asymmetric division.
In the one-cell embryo after fertilization, the male and female pronuclei meet in the posterior, but then move as complex along with the sperm associated centrosomes towards the center of the embryo (centration). The nuclear-centrosome complex also rotates 90°(nuclear rotation) to align the centrosomes on the polarized A/P axis (A/P) of the embryo During metaphase and anaphase, the entire spindle shifts posteriorly (spindle displacement) resulting in an unequal cleavage such that the anterior daughter AB cell is slightly larger than P1. Similar nuclear rotation and spindle displacement movements occur in the P1 cell.
copy021810 FM125 L4440 26hrsGREEN
A transgenic embryo expressing GFP::alpha-tubulin & mCherry::Histone H2B undergoing its first mitotic division. The asymmetric movements of centration, nuclear rotation and spindle displacement are shown. Imaged by spinning disc confocal. Espiritu et. al, Dev Bio, 2012.
The forces that mediate nuclear and spindle movements are generated by a conserved complex consisting of heterotrimeric Ga subunits and their activators (GPR and LIN-5 in C. elegans), which recruit the microtubule motor dynein to generate the cortical forces that pull on astral microtubules. Our studies identified LET-99, a DEPDC1 family protein, as a key regulator of the spindle positioning machinery. The highest levels of cortical LET-99 protein are restricted to a posterior lateral band through inhibition by PAR-3 at the anterior and PAR-1 at the posterior.
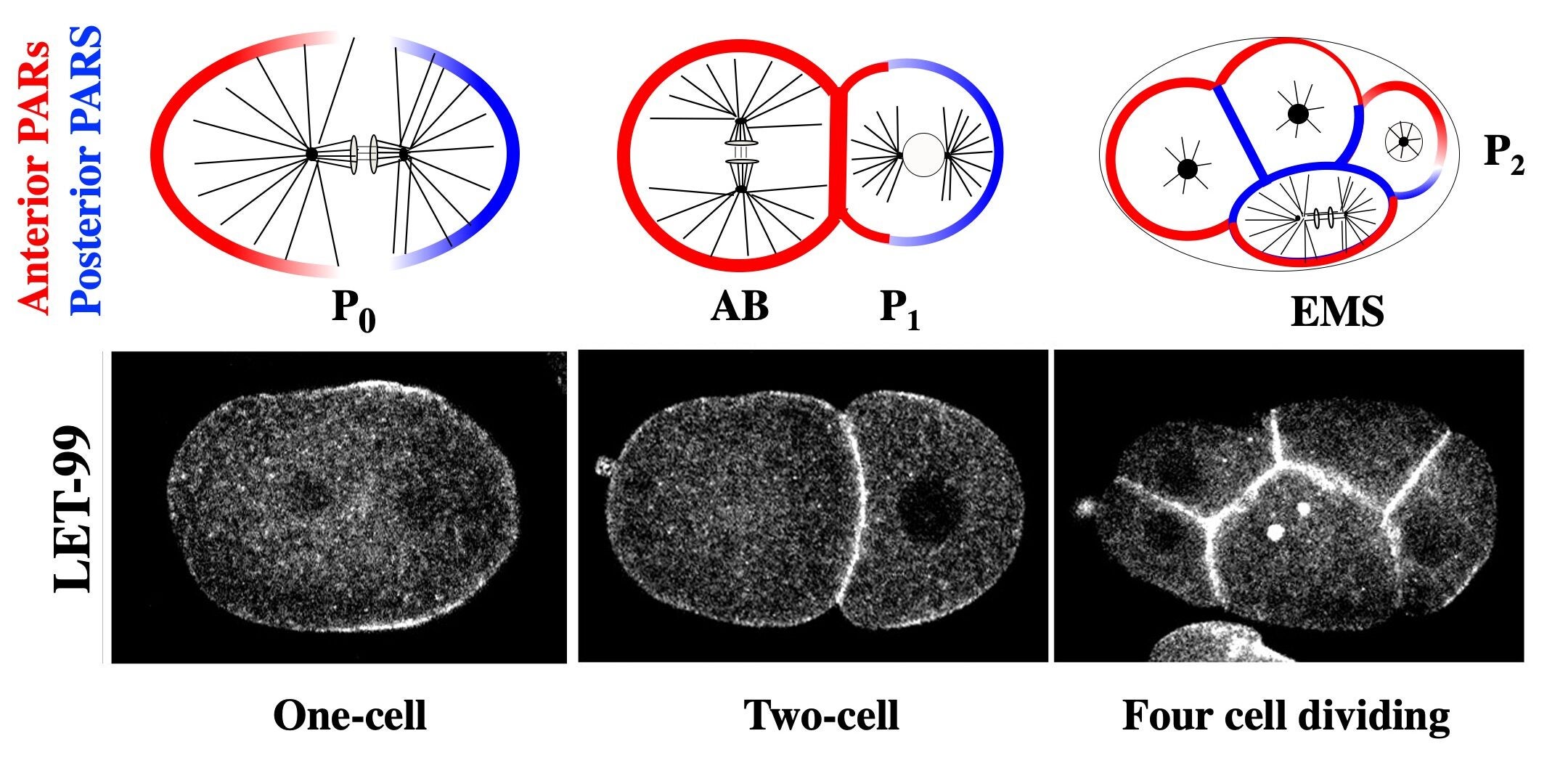
The anterior and posterior PARs are localized to opposite domains in the asymmetrically dividing P cells of the early embryo. LET-99 localizes to a posterior lateral band in those cells.
LET-99 in turn inhibits the cortical localization of GPR/LIN-5 in the band region, resulting in the asymmetric cortical accumulation of GPR/LIN-5 at the posterior-most cortex during spindle displacement. Significantly, we also showed that GPR-1/2 and LIN-5 are asymmetrically localized by LET-99 and the PARs to the opposite anterior cortex during the time of anteriorly directed nuclear rotation. These results provide the basis for our working model that the inhibition of G protein signaling in the LET-99 band region leads to three cortical force generation domains in the embryo (anterior, posterior lateral band and posterior); laser ablation studies that map the cortical forces support this model. We are currently dissecting the molecular mechanisms by which the PAR proteins regulate LET-99, and how LET-99 in turns regulates GPR localization.

Model for spindle positioning. Gα/GPR/LIN-5 are required for forces that pull on microtubules from the cortex (+), whereas LET-99 inhibits force. LET-99 and GPR/LIN-5 localize to the entire cortex, but only the regions with highest levels are indicated for simplicity. (a) Nuclear rotation & centration. (b) Spindle displacement . (c) Zoom of dashed area in (b). Espiritu & Rose, eLS, 2013
LET-99 during cytokinesis
Once the spindle is positioned, it signals the formation of the cytokinetic furrow to physically separate the cell. Timely formation of this furrow in the correct place is thus essential for an asymmetric division. The LET-99 band persists and becomes refined to the region of the ingressing cytokinetic furrow and work from other labs implicated LET-99 in furrow formation.
As in many systems, in the C. elegans one-cell embryo, both the astral and central spindle microtubules contribute to the localization and activation of the small G protein RhoA and thus contractile ring components at the equator. We have found that either the central spindle or astral microtubules can localize LET-99 to the furrow even in the absence of PAR proteins.
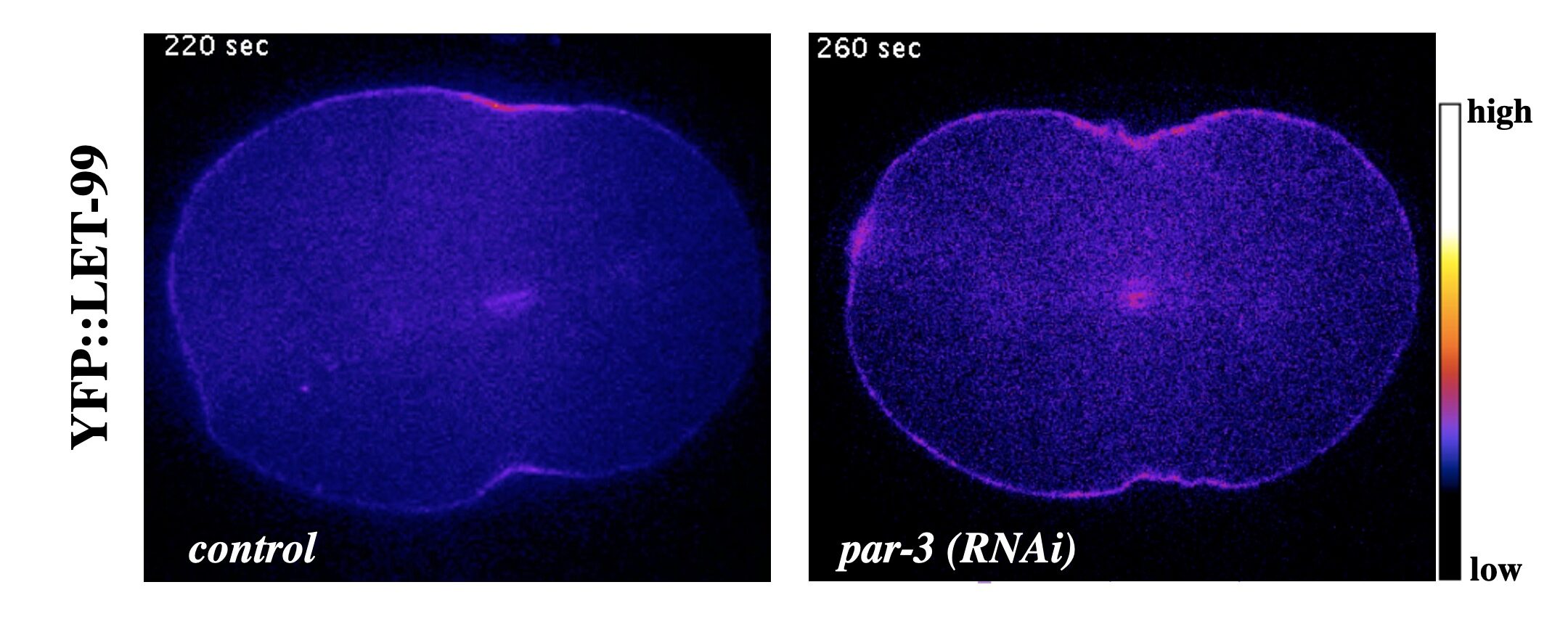
YFP::LET-99 localizes to the cleavage furrow during cytokinesis in wild type embryos and in polarity mutants
At this time and location, LET-99 promotes the proper timing of furrow ingression and the organization or enrichment of non-muscle myosin (NMY-2) at the equator. This role of LET-99 is genetically distinct from its role in spindle positioning, and occurs in both symmetrically and asymmetrically dividing cells. Our recent data suggests LET-99 antagonizes the formation of branched actin at the cortex, which is known to be inhibitory for furrowing. LET-99 is also required for overall cortical stability and with other data these results suggest that LET-99 directly or indirectly regulates the acto-myosin cytoskeleton in addition to the spindle positioning machinery.
Reestablishing polarity
How is the pattern of PAR domains reestablished for the next division? The mechanisms for establishing polarity in one-cell C. elegans embryo are well studied. After fertilization, a cue from the sperm associated centrosome results in an acto-myosin dependent movement that moves the anterior PARs away from the forming posterior end. However, it has been unknown how the P1 cell polarizes. We recently found that there is a novel early pathway dependent on the posterior PAR-1 kinase and inherited cytoplasmic polarity, and a late pathway that resembles symmetry breaking in the one-cell embryo.
LET-99 and EMS spindle positioning
The asymmetric division of the P1 cell results in daughter cells called EMS and P2 at the four-cell stage. The germline precursor P2 cell divides asymmetrically again, in response to PAR cues. However, the asymmetric division of the EMS cell is regulated by two partially redundant signals from P2 to EMS, which result in localization of cell fate determinants and alignment of the mitotic spindle along the anterior/posterior axis. Proper execution of this division is critical for generation of the endoderm (E, intestine) precursor.
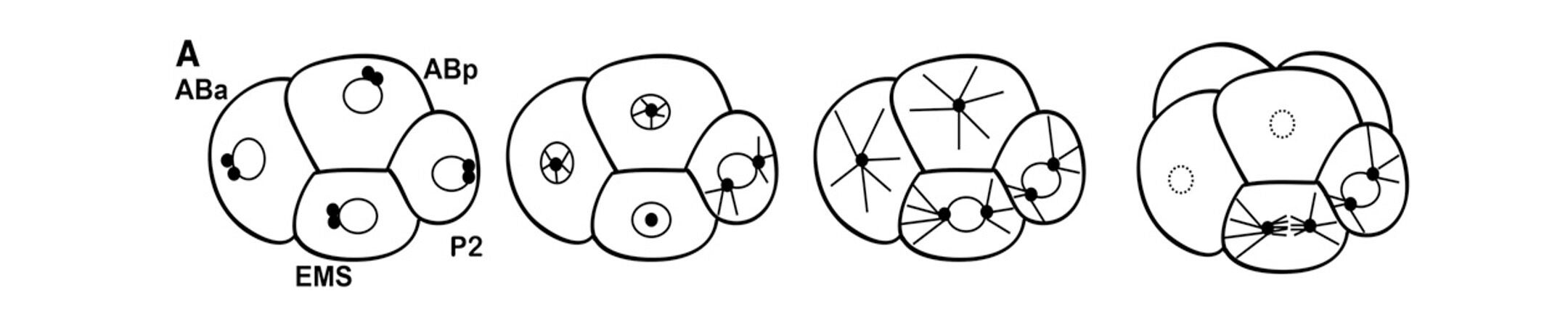
The centrosomes in the EMS cell migrate onto the left/right axis (in and out of the plane of view). The nuclear-centrosome complex rotates on to the anterior-posterior axis of the cell and the spindle stays on this axis.
We are working toward a complete molecular model of how these signals are transduced and how they regulate the EMS cytoskeleton for spindle orientation. We have identified the kinases PAR-1 and PIG-1, as well as LET-99 and the dynein adaptor LIN-5, as being involved in this asymmetric division. More recently, we found that the branched actin regulator Rac is also required for the EMS division.
Dissecting the many roles of LET-99
In addition to be involved in spindle positioning and cytokinesis, LET-99 localizes to the chromosomes and to the cortex at meiosis, and to the chromosomes and spindle midzone in mitosis. Further LET-99 has also been implicated in affecting programmed cell death in the germline. LET-99 has three predicted domains: the DEP domain, which binds to the membrane in other proteins, a region with partial homology to RHO GAP domains (RGL), which may interact with small GTPase proteins, and a hydrophobic region near the C-terminus. We are carrying out a structure-function analysis of LET-99 to determine which domains of LET-99 are required for its localization at different times and places and for its role in these different pathways.
C. elegans OOC-5 as a model for torsin function at nuclear pores
In the course of our studies on genes involved in asymmetric division, we identified a gene, ooc-5, as being required for the re-establishment of PAR polarity during the second asymmetric division in the embryo, as well as for normal oogenesis in adult C. elegans. OOC-5 is one of three C. elegans homologs of human torsinA, which is defective in individuals with the neuromuscular disease early onset dystonia. Like torsin A, OOC-5 is localized to the lumen of the endoplasmic reticulum and nuclear envelope. Torsins are part of the AAA+ family of proteins and may use ATP hydrolysis to dissociate or unfold target proteins.

Schematic of the C. elegans germline. Note the single row of growing oocytes diakenesis). In ooc-5 mutants, a double or multiple row forms. From VanGompel et al., MBoC, 2015
Interestingly, other C. elegans researchers showed that depletion of the NPP-1 nucleoporin, a component of the nuclear pore complex (NPC), leads to an asymmetric division phenotype highly similar to that of ooc-5 mutants. We have found that multiple nucleoporins are mislocalized in ooc-5 mutants, and the nuclear import is compromised. We plan to examine how ooc-5 and the NPC are involved in polarity regulation in embryos.